Checks before uploading¶
Check #1. Never use the special characters in your samples and contrasts, as certain characters are reserved for programming purposes. If you need to connect multiple elements, use underscore, “_” instead.
We test regularly which characters are allowed or not, we will update this list accordingly. The characters below are strictly forbidden.
Input file |
Part |
Characters not allowed |
|---|---|---|
sample.csv |
column names |
@ , : empty spaces + |
sample.csv |
row names |
@ , : empty spaces |
sample.csv |
phenotype names |
@ , : |
contrasts.csv |
column names |
@ , : empty spaces + |
contrasts.csv |
contrast names |
@ , : |
Note
We cannot guarantee that special characters (+, -, *, /, %, etc..) and empty space will work in all Omics Playground modules, so we recommend substituting them with underscore ‘_’.
Check #2 Avoid starting your sample, phenotype or contrast names with special characters like (+, -, *, /, %, etc..). While the platform will accept these characters, they may be converted into a standard symbol like X. For example, if we label the sample.csv country column as %country, we will see X.country in some analysis in Omics Playground.
Check #3 Make sure you have the words ‘samples’, ‘counts’ and ‘contrasts’ in the corresponding filenames. For example, we accept experiment23_samples.csv, samples_experiment23.csv, but not experiment23.csv as sample input.
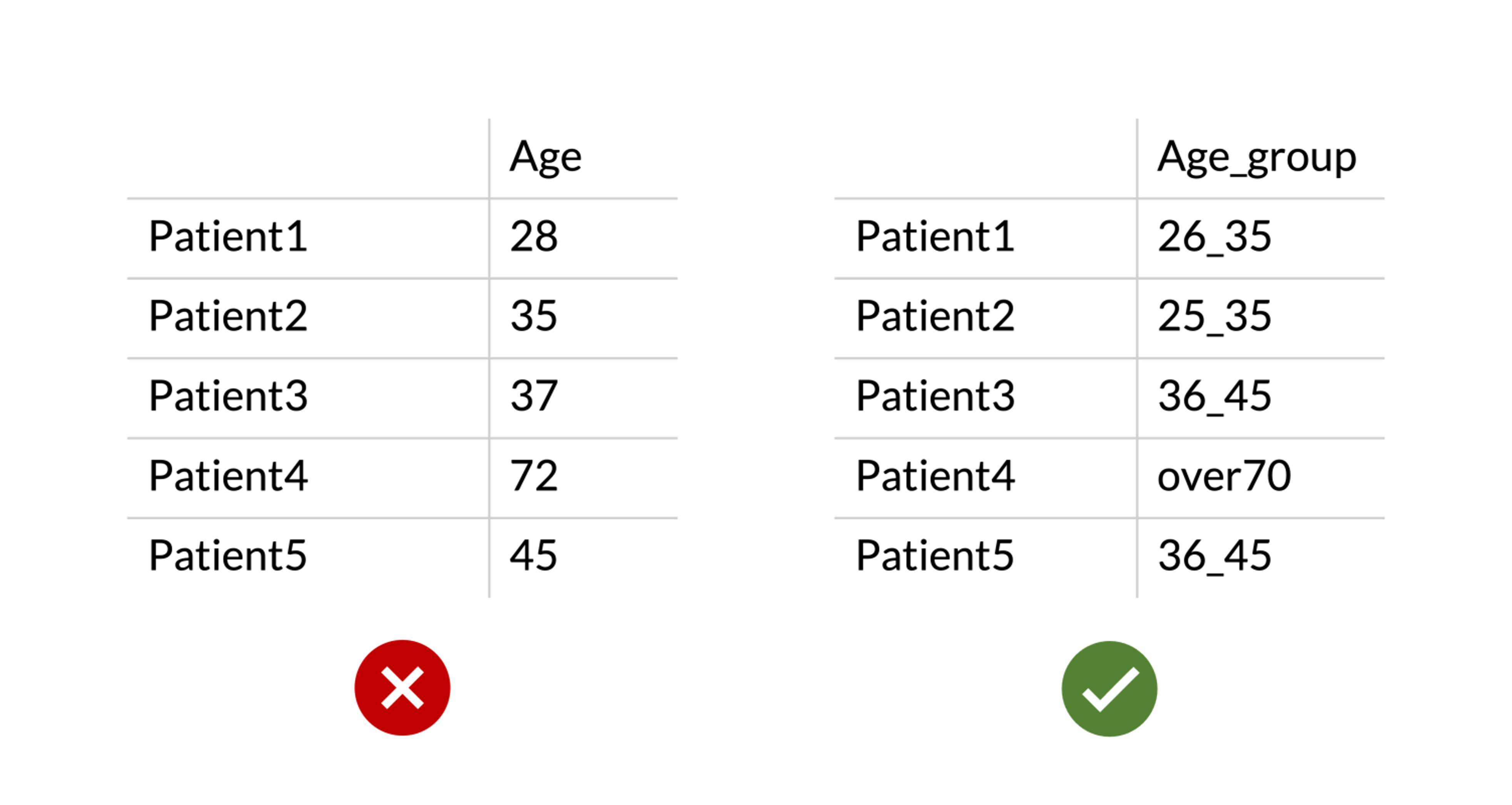
Check #4. Define intervals instead of using numeric phenotypes. The platform does not cope with continuous numeric variables for phenotypes yet. To avoid that, our coders added a filter that flags phenotypes names “Time” or “Age” as unacceptable. The same applies for other continuous variables, such as height, weight, length, etc.
Instead, you should cluster the various numeric values into definite intervals and then name them accordingly (e.g. “Age_groups”, “Time_intervals”, etc…)